-
PDF
- Split View
-
Views
-
Cite
Cite
Samy L. Habib, Besant K. Bhandari, Nahed Sadek, Sherry L. Abboud-Werner, Hanna E. Abboud, Novel mechanism of regulation of the DNA repair enzyme OGG1 in tuberin-deficient cells, Carcinogenesis, Volume 31, Issue 11, November 2010, Pages 2022–2030, https://doi.org/10.1093/carcin/bgq189
Close - Share Icon Share
Abstract
Tuberin (protein encodes by tuberous sclerosis complex 2, Tsc2 ) deficiency is associated with the decrease in the DNA repair enzyme 8-oxoG-DNA glycosylase (OGG1) in tumour kidney of tuberous sclerosis complex (TSC) patients. The purpose of this study was to elucidate the mechanisms by which tuberin regulates OGG1. The partial deficiency in tuberin expression that occurs in the renal proximal tubular cells and kidney cortex of the Eker rat is associated with decreased activator protein 4 (AP4) and OGG1 expression. A complete deficiency in tuberin is associated with loss of AP4 and OGG1 expression in kidney tumour from Eker rats and the accumulation of significant levels of 8-oxo-deoxyguanosine. Knockdown of tuberin expression in human renal epithelial cells (HEK293) with small interfering RNA (siRNA) also resulted in a marked decrease in the expression of AP4 and OGG1. In contrast, overexpression of tuberin in HEK293 cells increased the expression of AP4 and OGG1 proteins. Downregulation of AP4 expression using siRNA resulted in a significant decrease in the protein expression of OGG1. Immunoprecipitation studies show that AP4 is associated with tuberin in cells. Gel shift analysis and chromatin immunoprecipitation identified the transcription factor AP4 as a positive regulator of the OGG1 promoter. AP4 DNA-binding activity is significantly reduced in Tsc2−/− as compared with Tsc2+/+ cells. Transcriptional activity of the OGG1 promoter is also decreased in tuberin-null cells compared with wild-type cells. These data indicate a novel role for tuberin in the regulation of OGG1 through the transcription factor AP4. This regulation may be important in the pathogenesis of kidney tumours in patients with TSC disease.
Introduction
Tuberous sclerosis complex (TSC) is a genetic disorder associated with renal cell carcinoma ( 1 ). Several studies have identified the TSC2 protein tuberin as an Akt substrate providing another plausible signaling pathways to place mammalian target of rapamycin (mTOR) downstream from Akt ( 2–5 ). Phosphorylation by Akt results in phosphorylation and inactivation of Tsc2 by several mechanisms, including degradation of the Tsc1 – Tsc2 complex ( 6–11 ). Loss of heterozygosity at the TSC2 locus has been detected in Tsc -associated renal cell carcinoma as well as in the sporadic forms of these tumours ( 12 , 13 ). In the Eker rat where the Tsc2 gene is mutated, the incidence of renal cell tumours in gene carriers approaches 100% by 1 year of age ( 14–16 ). The precise mechanism by which tuberin deficiency predisposes to renal carcinogenesis is yet not known.
The mutagenic 8-oxo-deoxyguanosine (8-oxodG) is one of the prominent base lesions formed after the DNA has been damaged by oxidative stress and is repaired primarily via the DNA base excision repair pathway ( 17 , 18 ). 8-Oxoguanine-DNA glycosylase (OGG1) is the DNA repair enzyme that recognizes and excises 8-oxodG ( 19 , 20 ). The steady-state levels of 8-oxoguanine, which reflect the balance between the continuous generation and removal of this modification, are significantly higher in the liver of Ogg1 −/− mice compared with wild-type animals ( 21 ). Many human cancer cells exhibit somatic mutations in the OGG1 gene, whereas OGG1 is highly polymorphic ( 22–30 ). Loss of heterozygosity at the OGG1 allele is observed in 85% of human kidney clear cell carcinoma and loss of OGG1 function is a possible contributor to carcinogenesis in the kidney ( 31 ). In addition, genetic polymorphisms of OGG1 gene are associated with angiomyolipoma, a benign kidney tumour from TSC patients ( 32 ).
Transcription factor activator protein 4 (AP4) has recently been identified as a transcriptional repressor of HIV-1 ( 33 ). Viral and cellular genes are activated by AP4, which binds to the symmetrical DNA sequence CAGCTG in the promoter region of responsive genes ( 34 , 35 ). There is little information on the cellular and molecular mechanisms that regulate OGG1 . The constitutive expression of OGG1 protein in the kidneys of the Eker rat is decreased compared with wild-type rats ( 36 ). In addition, we have shown that tuberin haploinsufficiency is associated with the loss of OGG1 in rat kidney tumours ( 37 ). Our recent published data also demonstrate that tuberin regulates OGG1 expression and activity through the transcription factor NF-YA in renal epithelial cells ( 38 ) and in angiomyolipoma tissue of TSC patients ( 39 ).
Renal cell carcinoma arises from proximal tubular cells. The mechanism for the suppression of OGG1 in renal proximal tubular cells from these rats is not known. In the current study, we examined the role of tuberin in regulating AP4 that is involved in transcription of OGG1 . To extend the findings to known mechanistic pathways, we also demonstrate the role of tuberin in the modulation of OGG1 . Here, we report for the first time that OGG1 expression is positively regulated by AP4 in renal proximal tubular epithelial cells and kidney cortex of rat. The binding of AP4 to OGG1 promoter may provide a mechanism for enhancing the expression of the DNA repair gene, OGG1 to protect the cells against oxidative DNA damage.
Materials and methods
Cell cultures
Isolation and culture of primary proximal tubular cells.
Primary proximal tubular cells were isolated and cultured following the method of Glynne with minor modifications ( 40 ) from wild-type and Eker kidney rats. Renal cortical tissue was collected in cooled Hank’s balanced salt solution containing penicillin (50 U/ml), streptomycin (50 μg/ml) and amphotericin B (0.125 μg/ml). After the capsule was removed, the cortex was cut into small pieces and the tissue fragments were suspended in 1 mg/ml (in Hank’s balanced salt solution) of type II collagenase (Worthington Biochemical, Lakewood, NJ) and incubated for 1 h at 37°C. The cells were centrifuged (200 g , 5 min, 4°C) and seeded into 75 cm 2 tissue culture flasks that had been coated with collagen S. The cells were grown in serum-free medium (Dulbecco's modified Eagle's medium/F-12; glucose concentration, 17 mM) containing 15 mM N -2-hydroxyethylpiperazine- N ′-2-ethanesulfonic acid buffer, l -glutamine and pyridoxine hydrochloride. The medium was supplemented with epidermal growth factor (10 ng/ml), insulin (10 μg/ml), transferrin (5 μg/ml), selenium (5 ng/ml), hydrocortisone (36 ng/ml), triiodothyronine (4 pg/ml), penicillin (50 U/ml), streptomycin (50 μg/ml) and amphotericin B (0.125 μg/ml). The cells were incubated at 37°C in humidified 5% CO 2 in air.
Mouse embryonic fibroblasts cells.
Mouse embryonic fibroblasts derived from Tsc2−/− , Tsc2+/− and Tsc2+/+ embryos were generously provided by Dr D.J.Kwiatkowski (Harvard Medical School, Boston, MA). The cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat inactivated fetal bovine serum. Cells were used between passages 2 and 6.
Human embryonic kidney epithelial cells.
Human embryonic kidney epithelial cells (HEK293) were obtained from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. All cell lines were grown at 37°C in a humidified atmosphere of 5% CO 2 .
Animals
Male wild-type and Eker rats (wild-type, Tsc-2+/+ and mutant Tsc-2 +/− ) were purchased from a breeding colony maintained in house at the University of Texas M. D. Anderson Cancer Center, Smithville, TX. The animals were allowed food and water ad libitum during the experiments. Animals were euthanized at 12 months for nephrectomy. Kidneys were quickly removed and dissected longitudinally, half preserved in 10% formalin in phosphate-buffered saline (PBS), pH 7.4 for immunostaining and the remaining tissues were snap frozen in liquid nitrogen for biochemical analysis. Tumour tissues from Eker rats were dissected for immunostaining and biochemical analysis. The Institutional Animal Care and Use Committee of the University of Texas Health Science Center at San Antonio approved these animal studies.
Protein extraction and western blot analysis
Cell lysate and kidney tissue homogenates were prepared as described previously ( 37 , 38 ). Protein concentrations were determined with the Bradford assay ( 41 ) using bovine serum albumin as a standard. Western blot analysis was performed as described previously ( 38 ). Anti-tuberin (C-20) and GAPDH antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-AP4 antibody was purchased from CHEMICON International, Inc. (Temecula, CA). Anti-rabbit polyclonal OGG1 antibody was generously provided by Dr S.Mitra (University of Texas Medical Branch at Galveston, TX). All primary antibodies were prepared at 1:1000 dilutions in Tris-Buffered Saline-Tween-20. All Tris buffer saline-t secondary antibodies were prepared at 1:5000 dilutions in Tris-Buffered Saline-Tween-20. Enhanced chemiluminescence advance kit (Amersham, NJ) was used to identify the proteins on the western blots. Expression of each protein was quantified by densitometry using National Institutes of Health image 1.62 software and normalized to a loading control.
Histology of normal and tumour kidney
Formalin-fixed kidneys were embedded in paraffin 3 μm sections stained with haematoxylin and eosin for histological examination by light microscopy.
Immunostaining and high-performance liquid chromatography assays of 8-oxodG
DNA was isolated from kidney tissue of wild-type Eker rat as well as kidney tumour of Eker rats as described previously ( 36 ). Kidneys from wild-type and Eker rats were harvested at 12 months of age. Detection of deoxyguanosine (dG) and 8-oxodG was performed on DNA hydrolyzed with nuclease P1 and alkaline phosphatase as described previously and validated ( 36 ). Data were expressed as picomoles of 8-oxodG/dG Å ∼10 −5 in 90 μl of DNA hydrolysate. The amount of 8-oxodG was confirmed by immunostaining in kidney tissue of wild-type Eker rat and kidney tumour of Eker rat as described previously ( 32 ). A fluorescent labelling method was used as described previously ( 36 ) with minor modifications. Kidney sections were stained for 8-oxodG. Acetone-fixed frozen kidney sections (6 μm) were incubated with non-immune donkey IgG to block non-specific binding then incubated with mouse antibody against 8-oxo-dG (1:200) conjugated with fluorescein isothiocyanate (FITC) (Biodesign International, Saco, ME). The slides were reacted with Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA). In this assay, 8-oxo-dG was identified by the primary monoclonal antibody and FITC-conjugated secondary antibody. FITC green signals for 8-oxo-dG were detected using a filter with excitation range 450–490 nm. Sections were viewed and photographed using an Olympus Research microscope equipped for epifluorescence with excitation and band pass filters optimal for FITC. To demonstrate staining specificity, control kidney sections were stained without primary antibody.
Overexpression of tuberin
The HEK293 cells were grown to 30–50% confluence, made quiescent by serum deprivation for 24 h and then infected at room temperature for 1 h with adenovirus 6.01 carrying the Tsc2 gene ( 38 ). An adenovirus vector expressing protein (Adβ-GAL) was used as a control. The cells were then incubated for 48 h at 37°C in a humidified atmosphere of 5% CO 2 . Cells were washed twice with PBS buffer and western blot analysis was performed on the cell extracts as above using tuberin, AP4 and OGG1 antibodies.
siRNA experiments
HEK293 cells were grown in six-well plates. Prior to transfection, cells (30–50% confluent) were washed with PBS and media was replaced with 800 μl of OPTI-MEM I (Invitrogen, Carlsbad, CA). In parallel, 4 μl of oligofectamine (Invitrogen) were combined with 11 μl of OPTI-MEM I and incubated at room temperature for 10 min. SMART selected small interfering RNA (siRNA) duplexes of Tsc2 or AP4 with ‘UU’ overhangs and 5′ phosphate on the antisense strand. The siRNA specific for Tsc2 and AP4 were a mixture of four pooled duplexes. The Tsc2 siRNA kit was obtained from Dharmacon/Upstate, (Lake Placid, NY) and AP4 siRNA kit from Santa Cruz Biotechnology. According to the manufacturer, these siRNA efficiently blocks tuberin and AP4 expression by 50–70%. The indicated duplex of 1.5 μg were diluted into 180 μl of OPTI-MEM I, added to the Oligofectamine/OPTI-MEM I mixture and incubated at room temperature for 20 min ( 38 ). The siRNA complexes were then added to the cells. After incubation for 3–4 h in a 5% CO 2 incubator, 1 ml of fresh medium was added to a final serum concentration of 10%. Forty-eight hours after transfection, cells were harvested for western blot analysis. The control construct used in parallel experiments contains four pooled non-specific siRNA duplexes provided by with the kit ( 42 ).
Immunoprecipitation of AP4
AP4 was immunoprecipitated in Tsc2+/+ cell extracts with AP4 antibody using protein G-agarose according to manufacturer’s protocol (Santa Cruz Biotechnogies, Santa Cruz, CA). Cell lysates were immunoblotted with tuberin and AP4 antibodies.
Electrophoretic mobility shift assay
Nuclear proteins were extracted from Tsc2−/− , Tsc2+/− and Tsc2+/+ cells as described previously ( 38 ). The protein concentration of the nuclear extracts was determined using Bradford method ( 41 ). Electrophoretic mobility shift assay (EMSA)-binding reactions were incubated in a 20 μl final volume for 20 min at room temperature containing 5 μg of the nuclear extract, 20–30 fmol of the 5′ end-labelled double-stranded 21 bp oligonucleotide: 5′-AAGGCGAGCAGCTGGCAGAGA-3′, covering the region of the hOGG1 promoter from −156 to −176 (control), and 1 μl of poly (dI-dC)·(dI-dC). The super shift assays were performed by pre-incubating nuclear extracts with 1 and 5 μg of AP4 antibody (Santa Cruz Biotechnology) into the reaction. In addition, labelled AP4 oligonucleotide at −163/−168 nt region with Mut-1 (aAGCTG), Mut-2 (CAGaTG) or Mut-3 (aAGaTG) was incubated with nuclear extracts of Tsc2 +/+ cells. The reaction was carried out at room temperature for 30 min prior to adding the radiolabelled probe. Competition was performed in the presence of a 100-fold excess of the unlabelled oligonucleotides. The complexes were resolved using a 5% non-denaturing polyacrylamide gel. The gels were dried and exposed overnight at −70°C.
Chromatin immunoprecipitation assay
The assay was performed using a ChIP kit (Active Motif, Carlsbad, CA). Briefly, HEK293 cells grown in 100 mm dish were cross-linked using 1% formaldehyde ( 38 ). After washing twice with PBS, cells were resuspended in lysis buffer containing protease inhibitor cocktail. DNA was digested (5 min) to small fragments ranged from 100 to 500 bp by enzymatic method. The supernatant was then pre-cleared using protein G-agarose beads. After centrifugation, supernatant was incubated with either anti-AP4 antibody or control IgG for overnight. Incubation was further carried out with protein G-agarose beads for 2 h and beads were washed with buffers provided in the kit. The immunoprecipitated DNA was eluted from the beads with 1% sodium sulphate dodecyl and 1 M NaHCO 3 solution and reverse cross-linked using 5 M NaCl and RNase A at 65°C for overnight. Samples were then digested with proteinase K and DNA was purified using column method. Polymerase chain reaction (PCR) was performed on the purified DNA using hOGG1 promoter-specific primers forward (−209/−228) (5′-CCAGATGGAACTCGTTAGCG-3′) and reverse (−9/−29) (5′-CAGACCACAGCACCACCGGAA-3′).
Transcriptional activity of OGG1 promoter
The reporter plasmid consists of the OGG1 promoter with luciferase as reporter gene [courtesy of Dr Bioteux ( 43 )] was used to determine the transcriptional activity of OGG1 promoter in mouse embryonic fibroblasts cells ( 38 ). Plasmids were transfected into HEK293 cells using the LipofectAMINE and Plus Reagent™ method (Invitrogen, Life Technologies, Carlsbad, CA). The cells were grown in six-well plates to 60–70% confluence. Prior to transfection, cells were washed twice with PBS and media was replaced with 1 ml of OPTI-MEM I (Invitrogen). Pre-complex of the DNA with Plus Reagent™ in Opti-MEM was mixed and incubated at room temperature for 15 min. LipofectAMINE was added to the complex of DNA and Plus Reagent™ and incubated for 15 min at room temperature. DNA and Plus Reagent™–LipofectAMINE complexes was added to each well and incubated at 37°C with 5% CO 2 . After incubation for 4 h, 1 ml of fresh media with 20% serum was added to a final concentration of 10%. Cells were harvested after 48 h of transfection. Cells were washed twice with PBS and lysed in 0.2 ml of lysis buffer. Luciferase activity was determined using the Luciferase Reporter Assay System by a luminometer according to the manufacturer’s instructions (Promega, Madison, WI) and normalized to protein content.
Mutations in the AP4-binding site of the OGG1 promoter
Plasmids containing mutations at AP4-binding site in OGG1 promoter were constructed using PCR followed by cloning of PCR products into pGL3-basic vector (Promega, Madison, WI). Briefly, two forward primers containing a KpnI site at 5′ and sequences −171 to +74 of OGG1 promoter with mutations were synthesized: AP4 no mutation, 5′-CAGCTGGTACCGAGCAGCTGGCAGAGAGCCCA-GTGCCGGCCA-3′, AP4 Mut 1, 5′-CAGCTGGTACCGAG A AGCTGGCAGAGAGCCCAGTGCCGGCCA-3′, AP4 Mut 2, 5′-CAGCTGGTAACGAGCAG A TGGCAGAGAGCCCAGTGCCGGCCA-3′ and AP4 Mut 3, 5′-CAGCTGGTACCGAG A AG A TGGCAGAGAGCCCAGTGCCGGCCA-3′. (Nucleotide bases in bold are referring to the mutation sites). A reverse primer containing a HindIII site at 3′ sequences corresponding to pGL3 basic vector was also synthesized: 5′-CATGGTGGCTTTACCAACAGTACCGGAATG-3′. PCRs were performed using pR143 clone ( 44 ) as the template DNA with an appropriate set of primers, Taq polymerase and deoxynucleotide triphosphates, as described previously ( 44 ). The PCR products were digested with KpnI and HindIII and ligated into pGL3 basic vector using T4 ligase followed by transformation of DH5a cells (Invitrogen, Life Technologies). The mutations in the constructs were confirmed by DNA sequencing. The reporter plasmids with the OGG1 promoter driving firefly luciferase were used to determine the transcriptional activity of OGG1 promoter with and without mutations at AP4-binding site. Plasmids were transfected into mouse embryonic fibroblasts cells using the LipofectAMINE and Plus Reagent™ method (Invitrogen, Life Technologies) as described above.
Statistics
Data are presented as mean ± standard error. Statistical differences were determined using analysis of variance followed by Student–Dunnett’s (Experiment versus Control) test using 1 trial analysis. P -values <0.01 were considered statistically significant.
Results
Tuberin deficiency is associated with downregulation of AP4 and OGG1 expression in primary proximal tubular cells of the Eker rat
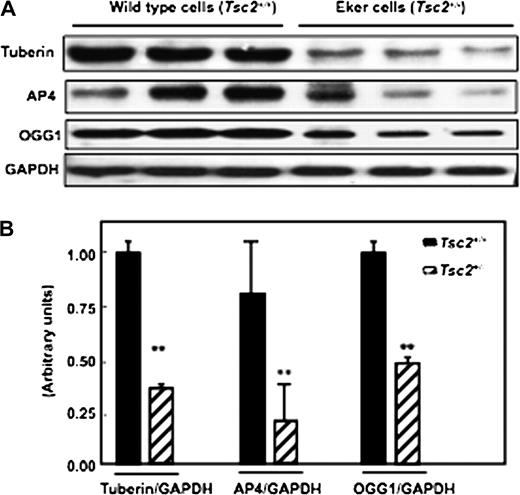
To determine if deficiency in tuberin is associated with decreased AP4 and OGG1, we performed western blot analysis on cell lysates of proximal tubular cells isolated from kidneys of Eker and wild-type rats. Decreased tuberin expression was associated with decreased AP4 and OGG1 expression in primary cells derived from normal Eker rats compared with wild-type cells ( Figure 1A&B ).
Partial deficiency in tuberin ( Tsc2+/− ) is associated with decreased AP4 and OGG1 expression in primary proximal tubular cells isolated from kidney cortex of Eker rat. ( A ) Immunoblot analysis of tuberin, OGG1 and AP4 in proximal tubular cells isolated from wild-type (lanes 1–3) and Eker (lanes 4–6) rats. Markedly decreased in AP4 and OGG1 expression was noted in proximal tubular cells isolated from Eker rat compared with proximal tubular cells isolated from wild-type rats. GAPDH was used as a loading control. ( B ) Histograms represent means (n = 3) ± SE. Significant difference from wild-type cells is indicated by ** P < 0.01.
Deficiency of tuberin is associated with the loss of AP4 and OGG1 expression in kidney tumour of Eker rat
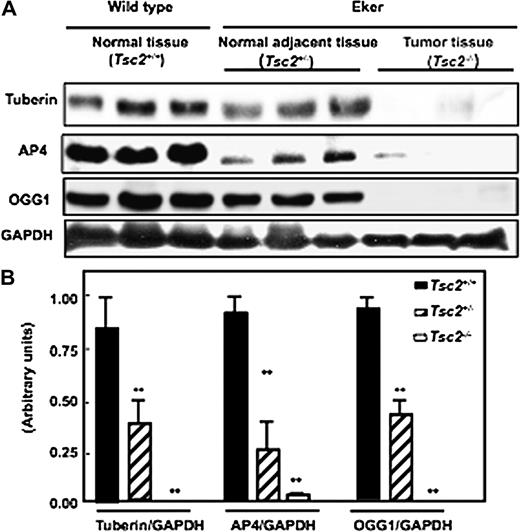
To determine if tuberin deficiency in kidney tumour of Eker rat is associated with downregulation of AP4 and OGG1 expression, kidney cortex homogenates from wild-type, normal and tumour of Eker rat were analysed by western blot. Tuberin expression in kidney tumours from Eker rats is significantly reduced compared with normal kidney tissues of Eker and wild-type rats. Tuberin deficiency is associated with the loss of AP4 and OGG1 expression in kidney tumour of Eker rat ( Figure 2A&B ). These data demonstrate that tuberin may be an important regulator of AP4 and OGG1 expression.
Deficiency of tuberin is associated with the loss of AP4 and OGG1 expression in kidney tumour of Eker rat. ( A ) Immunoblot analysis of tuberin, AP4 and OGG1 expression in kidney cortex of wild-type ( Tsc2+/+ ) (lanes 1–3), normal kidney of Eker rat ( Tsc2+/− ) (lanes 4–6) and tumour kidney ( Tsc2−/− ) (lanes 7–9) of Eker rat. Partial deficiency in tuberin is associated with decreased protein expression of AP4 and OGG1. Complete loss of tuberin expression is associated with abolished AP4 and OGG1 expression in kidney tumour tissues of Eker rat. GAPDH was used as loading control. ( B ) Histograms represent means ± SE of three animals. Significant difference from wild-type animals is indicated by ** P < 0.01.
Loss of OGG1 is associated with significant accumulation of 8-oxodG
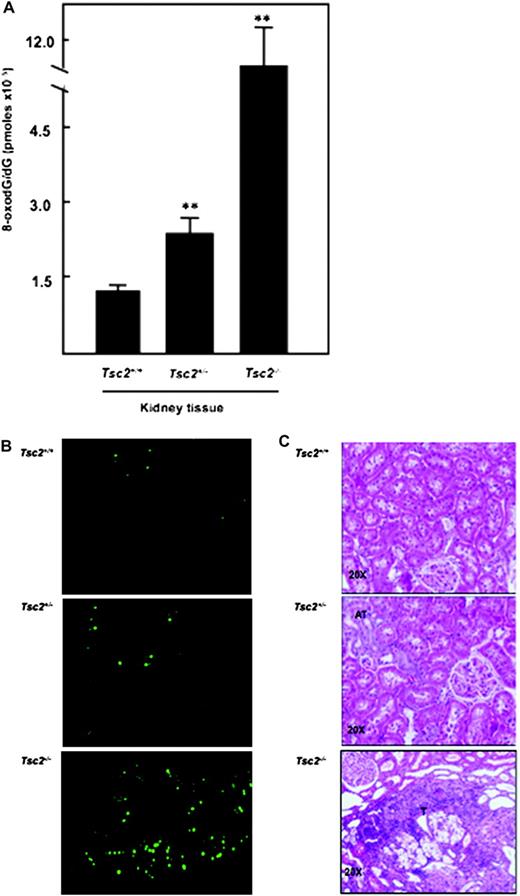
The basal levels of 8-oxodG as measured by high-performance liquid chromatography were 2-fold higher in kidney cortex of Eker rat compared with wild-type rat ( Figure 3 ). In addition, tumour kidney of Eker rat is associated with an 8-fold accumulation of 8-oxodG compared with normal kidney tissue from wild-type rat and 6-fold compared with kidney tissue of Eker rats ( Figure 3A ). Immunohistochemistry of 8-oxodG confirmed that significant accumulation of 8-oxodG in tumour kidney from Eker rat compared with normal kidney from Eker and wild-type rats ( Figure 3B ). The histological appearance of haematoxylin- and eosin-stained kidney sections of wild type showed normal tubular architecture of kidney. Tumours containing cells with clear cytoplasm and excentric in large nuclei of clear cell carcinoma type were appeared in the kidney section of Eker rat ( Figure 3C ).
Significant increase in 8-oxodG levels in kidney tumour of Eker rat. ( A ) DNA was extracted from normal kidney of wild-type and Eker rat as well as from kidney tumour of Eker rat. Detection of dG and 8-oxodG was performed by High Pressure Liquid Chromatography-Electrochemical analysis. Authentic standards of 8-oxodG and dG were analysed simultaneously. Standard curves for dG and 8-oxodG were prepared and quantitated by linear regression analyses. Histograms represent means ± SE of three animals per group. Significant difference between wild-type, Eker rat and tumour of Eker rat is indicated by ** P < 0.01. ( B ) These data confirmed by immunohistochemistry in kidney sections from wild-type and Eker rat as well as from kidney tumour of Eker rats. FITC for 8-oxodG (green color) was detected with excitation wavelengths at 450–490 nm. ( C ) Haematoxylin and eosin staining of kidney sections of wild type showed normal tubular architecture. Kidney sections of Eker rat show atypical tubular (AT) in dark color and tumors (T) containing cells with clear cytoplasm and excentric in large nuclei of clear cell carcinoma type.
Tuberin regulates the expression of AP4 and OGG1 in renal epithelial cells
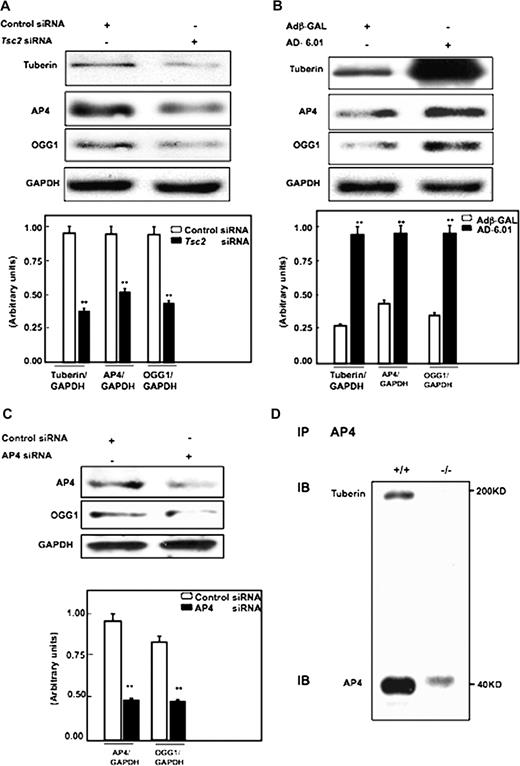
Our recent published data show that tuberin regulates OGG1 expression and activity through the transcription factor NF-YA in renal epithelial cells. To determine if tuberin also regulates OGG1 expression through the transcription factor AP4, tuberin was downregulated using duplex siRNA oligonucleotide complementary to TSC2 gene in human epithelial (HEK293) cells. Reduction of tuberin resulted in decreased AP4 and OGG1 expression compared with cells transfected with scrambled control oligonucleotides ( Figure 4A ). In contrast, overexpression of tuberin in HEK293 cells using an adenovirus expressing tuberin ( Ad-TSC2 ) results in an increase in AP4 and OGG1 expression, indicating that tuberin is an upstream regulator of AP4 and OGG1 ( Figure 4B ).
( A ) Tuberin regulates AP4 and OGG1 expression in human cells. Immunoblot analysis shows downregulation of tuberin with siRNA directed against Tsc2 is associated with decreased AP4 and OGG1 expression in human renal embryonic epithelial cells (HEK293). ( B ) Overexpression of tuberin increases AP4 and OGG1 expression. HEK293 cells were infected with adenovirus 6.01 expressing tuberin complementary DNA. An adenovirus vector expressing protein (Adβ-GAL) was used as a control. Overexpression of tuberin is associated with increased AP4 and OGG1 expression. GAPDH was used as a loading control. ( C ) Downregulation of AP4 results in significant decrease in OGG1 protein expression. Histograms represent means (n = 3) ± SE. Significant difference from cells treated with control siRNA or infected with Adβ-GAL is indicated by ** P < 0.01. ( D ) AP4 is associated with tuberin. Protein extracts from Tsc2+/+ cells were immunoprecipitated (IP) with AP4 antibody and immunoblotted (IB) with tuberin and AP4 antibody.
Downregulation of AP4 results in decrease OGG1 protein expression
To determine if AP4 regulates OGG1 expression, AP4 was reduced using duplex siRNA oligonucleotide complementary to AP4 gene in human epithelial (HEK293) cells. Downregulation of AP4 resulted in a decrease in OGG1 expression compared with cells transfected with scrambled control oligonucleotides ( Figure 4C ).
AP4 is associated with tuberin in cells
To investigate potential mechanisms by which tuberin regulates AP4, we performed co-immunoprecipitation analysis. Cell extracts from otherwise identical Tsc2+/+ or Tsc2−/− cells were immunoprecipitated with AP4 ( Figure 4D ) antibody, followed by immunoblotting with tuberin and AP4 antibodies. Our data show that AP4 co-precipitates with tuberin suggesting that AP4 is associated with tuberin in cells.
Tuberin deficiency decreases AP4 binding to OGG1 promoter
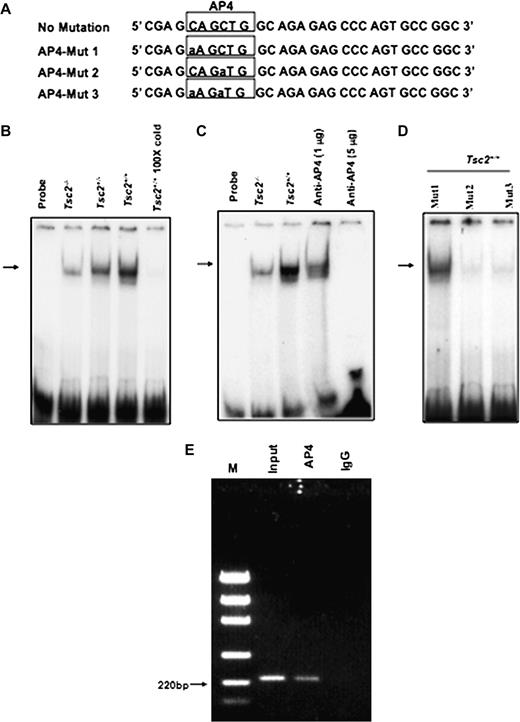
To further investigate the mechanism by which tuberin deficiency affects OGG1 expression, EMSAs were performed to determine if AP4 binds to DNA-binding site in the OGG1 promoter ( Figure 5A ). Binding of AP4 in Tsc2+/− , Tsc2+/− or Tsc2+/+ cells was analysed by EMSA using labelled oligonucleotide corresponding to the OGG1 promoter region between −156 and −176, which contains a consensus AP4-binding site ( Figure 5B ). Specific AP4 protein–DNA complexes were observed in the nuclear extracts prepared from Tsc2−/− , Tsc2+/− or Tsc2+/+ cells ( Figure 5B , lanes 2–4) with higher activity in Tsc2+/+ cells compared with Tsc2+/− and Tsc2−/− cells ( Figure 5B , lanes 2 and 4). Furthermore, the protein–DNA complex was competed effectively when nuclear extracts were incubated with 100-fold excess of the specific AP4 oligonucleotide ( Figure 5B , lane 5). The protein–DNA complex was also competed when nuclear extracts were incubated with AP4-specific antibodies ( Figure 5C , lanes 4–5). Furthermore, significant decrease of the protein–DNA complex was detected when Tsc2+/+ nuclear extracts were incubated with Mut-2 (CAGaTG) or Mut-3 (aAGaTG) but not Mut1 (aAGCTG) oligonucleotides of AP4 ( Figure 5D ). These data suggest that the −163/−168 nt region is a potentially important site for the regulation of OGG1 by AP4.
EMSA with the hOGG1 promoter region containing the AP4 motif in wild-type (TSC2 +/+ ) and TSC2-deficient (TSC2 −/−) cells. ( A ) Double-stranded oligonucleotide AP4 spanning the region of the hOGG1 promoter from −176 nt to −156 nt is shown. Consensus DNA binding for AP4 is underlined. ( B ) A 21 bp end-labelled double-stranded oligonucleotide spanning the region of the hOGG1 promoter from −176 nt to −156 nt was incubated without (lane 1) or with nuclear extracts isolated from Tsc2−/− (lane 2), Tsc2−/+ (lane 3) or Tsc2+/+ (lane 4) cells. Competition was performed in the presence of a 100-fold excess of unlabelled oligonucleotide (lane 5). C. Labelled AP4 oligonucleotide was incubated without nuclear extracts (lane 1) or with nuclear extracts from Tsc2−/− (lane 3) and Tsc2+/+ (lanes 4–6) cell lines as in (B). The DNA–AP4 protein complex was competed with either 1 or 5 μg AP4 antibody (lanes 4 and 5). The reaction mixtures were run on to 6% polyacrylamide gels and protein–DNA complexes shown by an arrow were visualized by autoradiography. ( D ) Labelled oligonucleotide at −163/−168 nt region Mut-1 (aAGCTG), Mut-2 (CAGaTG) or Mut-3 (aAGaTG) of AP4 were incubated with nuclear extracts of Tsc2+/+ cells. Mut-2 and Mut-3 show significant decrease of the protein–DNA complex compared with Mut-1. ( E ) ChIP assay showing that AP4 is involved in regulation of OGG1 transcription. Immunoprecipitation from HEK293 cell lysates was performed using an AP4 antibody or a non-specific goat IgG. The protein DNA complexes were denatured, and DNA was purified followed by a PCR using OGG1 promoter primers. The PCR was performed on the input (clarified chromatin from genomic DNA), as well as on the DNA immunoprecipitated using anti- AP4 and nonspecific goat IgG. PCR products were analysed by electrophoresis on a 1.5% agarose gel, and the size of the specific PCR amplified OGG1 promoter fragment (220 bp) was determined using the DNA markers (M). The results shown are representative of three experiments (n = 3).
To confirm if AP4 directly binds the DNA-binding region CAGCTG in the human OGG1 promoter, we performed chromatin immunoprecipitation (ChIP) assay using chromatin extracted from human 293 cells. The anti-AP4 antibody, but not the control IgG antibody, precipitated the OGG1 promoter fragment (220 bp) containing binding sites for AP4 region at −163/−168 nt ( Figure 5E ). These data show that AP4 does indeed bind to the OGG1 promoter at a CAGCTG site located in the region −163/−168 nt.
Mutation of the AP4-binding site inhibits the activity of OGG1 promoter
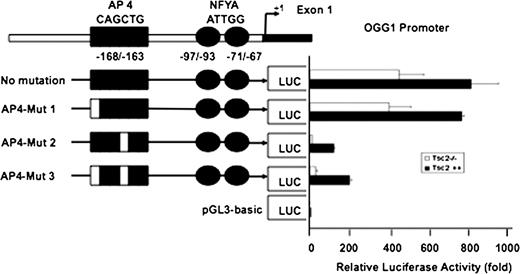
To determine the role of the AP4-binding site on OGG1 promoter activity, cells were transfected with AP4 mutant and wild-type reporter plasmids ( Figure 5A ). Mutation of the first ‘C’ of the AP4-binding site (CAGCTG to AAGCTG, AP4-Mut 1) had no effect on the OGG1 promoter activity. Mutation of the second C (CAGCTG to CAGATG, AP4-Mut 2) significantly inhibited OGG1 promoter activity ( Figure 6 ) as did mutation of both Cs of the AP4 motif from CAGCTG to AAGATG (AP4-Mut 3) ( Figure 6 ). These results strongly suggest that the potential AP4-binding site at −163/168 is important for the OGG1 promoter activity.
Luciferase activity of OGG1 promoter in Tsc2+/+ and Tsc2−/− cells. The reporter plasmid containing OGG1 promoter (−171/+74) that drive the expression of the luciferase gene was transfected into the Tsc2−/− and Tsc2+/+ cells. Forty-eight hours after transfection, luciferase activity was determined using the Luciferase Reporter Assay System. pGL3 basic vector was used as a control to normalize luciferase activity (1-fold). Mut-2 and Mut-3 show significant decrease of the OGG1 promoter activity compared with Mut-1. Histograms represent means ± SE from three experiments.
Discussion
These data comprise the first report to provide that tuberin regulates OGG1 expression through the transcription factors AP4 in cultured cells and in rat kidney cortex. Several approaches were utilized to study the potential role of tuberin in the regulation of OGG1 . First, we showed that decreased tuberin expression is associated with decreased AP4 and OGG1 expression in renal proximal tubular cells isolated from Eker rat as compared with wild-type cells. Second, decreased tuberin expression is associated also with decreased AP4 and OGG1 expression as well as significantly increased accumulation of 8-oxodG in kidney cortex of Eker compared with wild-type rat. In the kidney tumours of Eker rat, loss of tuberin is associated with loss of AP4 and OGG1 expression and an 8-fold increase in 8-oxodG compared with kidney tissue of wild-type rat. Third, we show that reduction of tuberin expression in human epithelial cells (293) using siRNA resulted in a marked decrease in the expression of OGG1 and AP4. Fourth, introduction of Tsc2 complementary DNA into tuberin-deficient cells restored OGG1 and AP4 expression. In addition, reduction of AP4 using siRNA results in significant decrease in OGG1 protein expression. Our data also show that AP4 is associated with tuberin in cells. We investigated the mechanism by which tuberin regulates OGG1 by gel shift analyses and ChIP and identified the transcription factor AP4 is a regulator of OGG1 promoter. Furthermore, we found that the DNA-binding activity of AP4 was significantly reduced in Tsc2−/− compared with Tsc2+/+ cells. In addition, the transcriptional activity of the OGG1 promoter was also decreased in Tsc2−/− cells. We find also that mutation(s) in AP4-binding region significantly inhibited OGG1 promoter activity suggesting that AP4 in an important enhancer of OGG1 promoter activity.
Tuberin expression in kidney from Eker rats is significantly reduced compared with expression in normal kidney tissues from wild-type littermates ( 37 ). In the current study, we show that loss of tuberin was associated with abolished AP4 and OGG1 expression as well as accumulation of significant levels of 8-oxodG in the kidney tumours of Eker rats. Downregulation of tuberin using siRNA directed specific for the Tsc2 gene in 293 human renal epithelial cells was associated with decreased in AP4 and OGG1 expression. In addition, overexpression of tuberin by adenovirus ( Ad - Tsc2 ) shows an increased in AP4 and OGG1 expression, indicating that tuberin is upstream of AP4 and OGG1. The immunoprecipitation analysis data of AP4 show that tuberin co-precipitates with AP4 indicating that AP4 is associated with tuberin in cells.
AP4 is an ubiquitously expressed transcription factor. It binds to the consensus sequence ‘CAGCTG’ ( 34 ). AP4 recognizes specific binding sites in the promoter as well as enhancer sequences of viral genes ( 34 ). The enhancer activity is abolished by mutations in these regions that also affect binding of AP4 in vitro ( 34 ). AP4 has recently been identified as a transcriptional repressor of HIV-1 ( 35 ). In the present study, we screened for enhancer-binding proteins to explore the molecular mechanisms that regulate OGG1 gene expression. We found that the OGG1 promoter contains a putative binding site for AP4 close to 3′ of the promoter region (−163/−168 nt).
Further investigation to test the role of tuberin in the regulation OGG1 by AP4 was performed by EMSA and ChIP analysis. Our data showed that AP4 binding was significantly lower in Tsc2−/− and Tsc2+/− cells compared with Tsc2+/+ cells. In addition, incubation of nuclear extracts with AP4-specific antibodies was competed the protein–DNA complex demonstrate that the Tsc2+/+ cells contain a protein, AP4, that binds specifically to a region in the OGG1 promoter. In addition, mutations of oligonucleotide (CAGaTG and aAGaTG) at −163/−168 nt region of AP4 show significant decrease in the protein–DNA complex. ChIP analysis further provided confirmation that Ap4 binds the OGG1 promoter at sites located at −163/−168 nt region. Collectively, the data indicate decreased binding of AP4 to the OGG1 promoter in tuberin-deficient cells. In addition, the mutations in consensus sequence of AP4 motif (CAGCTG to CAGATG) significantly inhibited OGG1 promoter activity strongly suggest that CAGCTG, at which AP4 is purported to bind to OGG1 promoter, is important for the OGG1 promoter activity.
In summary, these data indicate that tuberin regulates the OGG1 and this effect is at least partially through the transcription factor AP4. The transcription factor AP4 is an important regulator of the OGG1 promoter in the cell cultured and kidney tissue. Accumulation of significant levels of 8-oxodG in tumour kidney of Eker rat indicate that tuberin plays a significant role in protecting the cells from oxidative DNA damage. It is probably that loss of tuberin results in loss of AP4 and OGG1 expression and function in that may expose the cells to further genetic alterations and leading to accumulation of mismatched DNA base lesions, a form of genomic instability that if not repaired could accelerate further genetic alterations leading to the full-blown tumour phenotype in kidneys. These data indicate a novel role for tuberin in the regulation of DNA repair pathway through the transcription factor AP4 and provide a potential mechanism of Tsc2 and OGG1 in progression of kidney tumours.
Funding
American Diabetes Association Research Grant; Merit Review Award and New Investigator Award from the South Texas Veterans Healthcare System to S.L.H.
Abbreviations
- AP4
activator protein 4
- ChIP
chromatin immunoprecipitation
- dG
deoxyguanosine
- EMSA
electrophoretic mobility shift assay
- FITC
fluorescein isothiocyanate
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- 8-oxodG
8-oxo-deoxyguanosine
- OGG1
8-oxoG-DNA glycosylase
- siRNA
small interfering RNA
- TSC
tuberous sclerosis complex
The authors would like to acknowledge Dr Daniel J.Riley and Dr Anthony Valente for helpful discussions and for critical reading of the manuscript, Dr P.Radicella at Radiobiologie Moleculaire et Cellulaire, France, for providing the OGG1 promoter construct and Dr S.Mitra at the University of Texas M. D. Anderson and Sealy Center for Molecular Science, Galveston, TX for providing the OGG1 antibody. N.S. is a recipient of the Research Fellowship Award from The National Organization for Research, Egypt.
Conflict of Interest Statement: None declared.
References
- transcription, genetic
- immunoprecipitation
- mutation
- chromatin
- deoxyguanosine
- dna
- dna ligases
- down-regulation
- gel
- cortex of kidney
- kidney neoplasms
- tuberous sclerosis
- kidney
- neoplasms
- rats
- transcription factor
- transcriptional activation
- epithelial cells
- rna, small interfering
- protein overexpression
- hek293 cells